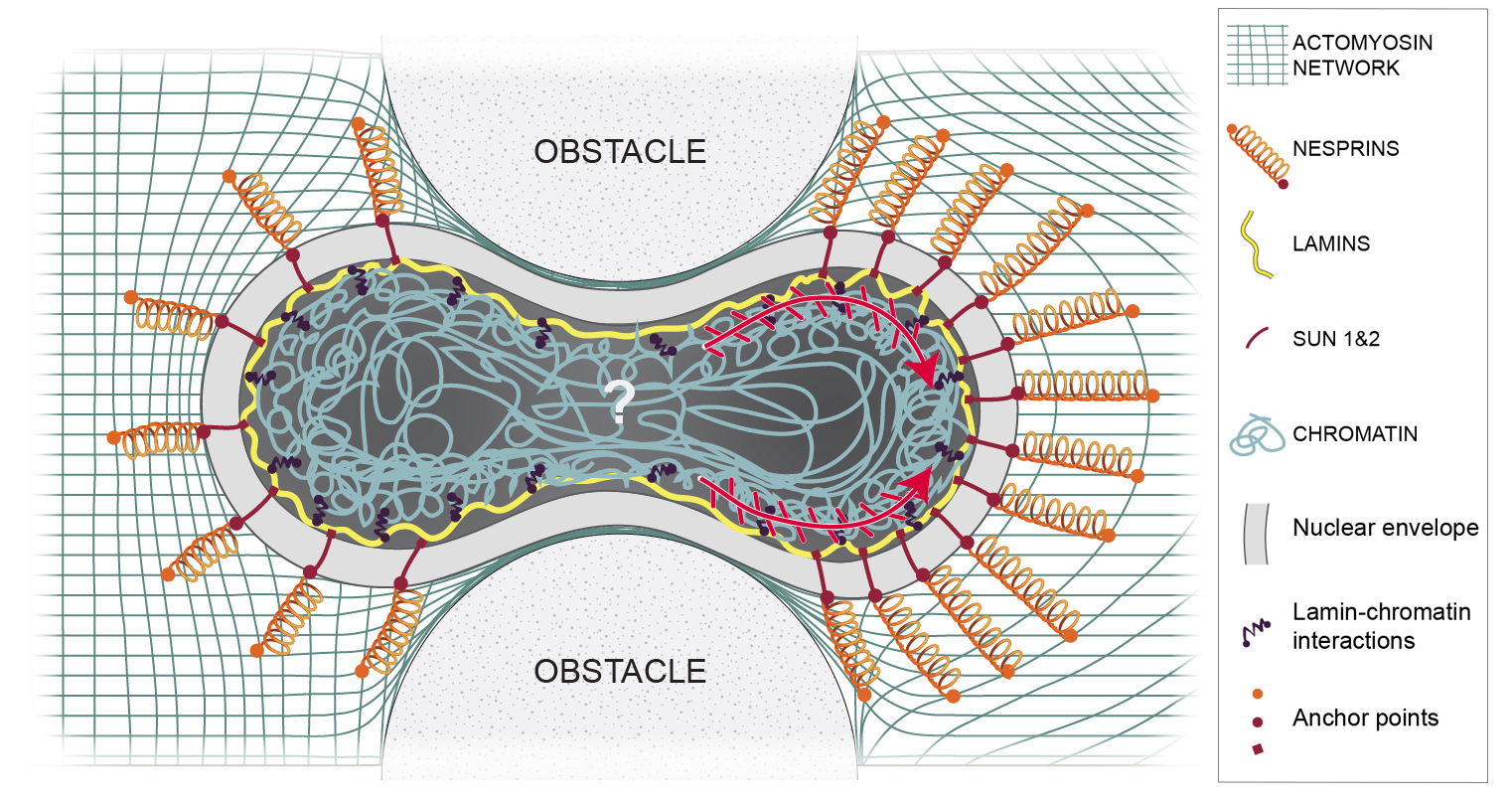
To understand the generic laws of nuclear “actuation” during confined cell movements, we use two complementary experimental approaches: microfluidic devices to observe individual migrating cells in a controlled environment, and a model organism, the worm Caenorhabditis elegans, to observe cell migration and invasion in a physiological context. We characterize nuclear deformation and nuclear protein dynamics along with modifications of the genetic material and the dynamics of the cytoskeleton during the movement and positioning of cells in these complex environments. We tackle the question at different scales:
– molecular, with simplified in vitro systems consisting of nuclei/nucleus-like objects and purified proteins that dynamically assemble to reproduce in vivo forces
– mesoscopic, with coarse-grained approaches where the dynamics of molecular distributions of nuclear proteins during cell motility are quantified and modeled,
– tissue, by studying nuclear and cytoskeletal dynamics in motile and invasive cells in vivo in the nematode C. elegans, in comparison with mammalian cells in microfluidic devices.
- The personal webpages of the members of the team can be accessed on the directory.