In biology, many RNA molecules act as sophisticated microscopic machines. Among them, riboswitches function as tiny biological sensors, changing their 3D shape upon binding to a specific metabolite. This shape-change acts as a switch, often turning a downstream gene « on » or « off ». The ability to design artificial switches from scratch would hold immense promise for synthetic biology, drug design, and new diagnostic tools. However, designing a sequence that can stably fold into two different shapes and switch between them is an extremely difficult challenge.
A collaboration involving CNRS researchers at École Normale Supérieure Paris, Université Paris Cité, École Polytechnique, and the Institut de Physique Théorique, have successfully used a machine learning approach to design entirely new, functional RNA switches. The team applied a model known as Restricted Boltzmann machines (RBM), to learn the « design rules » of the aptamer (1) domain of a natural riboswitch family. By training the RBM on thousands of natural sequences, the model learned to capture the complex interaction patterns leading to the formation of secondary and tertiary contacts (2) and essential for the structural switch, which simpler models miss.
The trained RBM was then used as a « generator » to create 476 novel RNA sequences, some differing by up to 40% from known natural sequences. The team synthesized and tested these artificial sequences using high-throughput chemical probing (SHAPE and DMS). The results were a remarkable success: approximately one-third of the high-scoring designed sequences functioned as effective switches, changing their conformation in response to the target metabolite (SAM) just as their natural counterparts do.
This work, recently published in Nature Communications, is a significant leap forward in the rational design of allosteric biomolecules. It demonstrates how generative models, born from the intersection of statistical physics and AI, can not only understand the complex language of biology but also use it to design new, functional molecules, paving the way for custom-designed molecular tools.
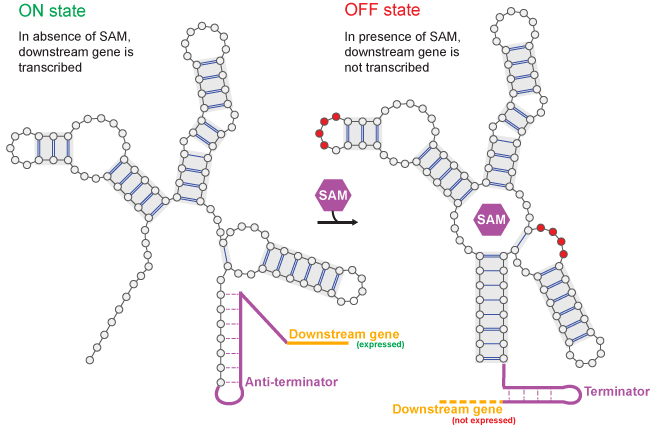
A riboswitch acts as a biological sensor, changing its 3D shape from an « ON » state (left) to an « OFF » state (right) when it binds a metabolite (SAM, purple hexagon). Researchers at the CNRS used a machine learning model to learn the design rules for this switch, enabling them to create entirely new, functional artificial molecules from scratch.
1. The aptamer domain is the part of the riboswitch responsible for recognizing and binding a specific metabolite.
2. Secondary and tertiary contacts refer to particular interactions between sites distant along the sequence that become close in 3D space when the RNA molecule is folded, such as pseudoknots.
More:
Article in Nature Communications
Affiliation author:
Laboratoire de physique de L’École normale supérieure (LPENS, ENS Paris/CNRS/Sorbonne Université/Université de Paris)
Corresponding author : Simona Cocco
Communication contact: Communication team